Somatic Mutational Rate (SMR)
Highlights:
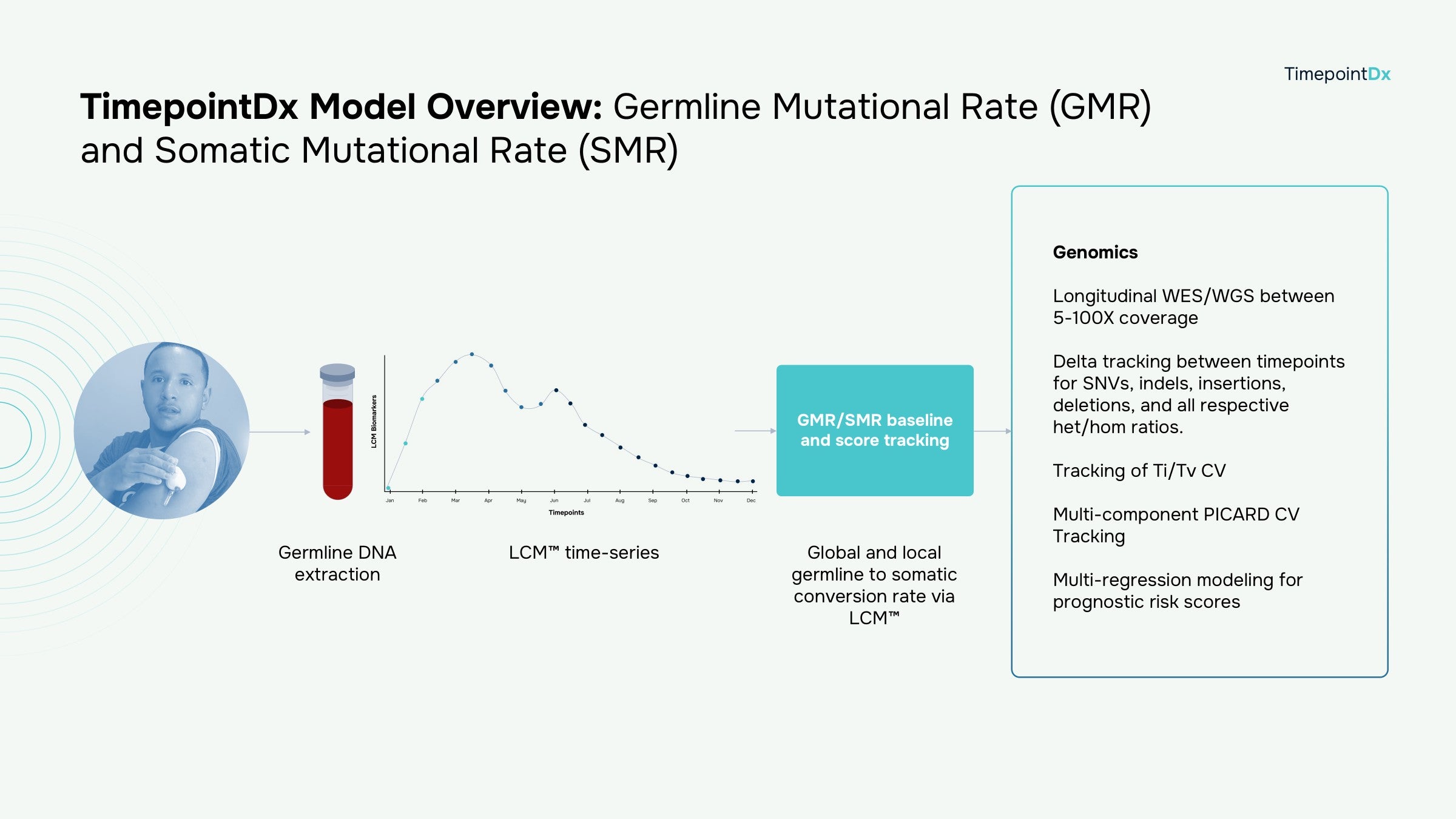
- Germline mutations increase the likelihood that an individual will develop disease. Somatic Mutation Rate (SMR) identifies the rate of change over a lifelong longitudinal assessment as compared to germline (constitutional baseline) and all subsequent somatic timepoints.
- SMR shows a significantly higher rate of change as compared to the germline mutational rate (GMR). Successive timepoints via TimepointDx LCM™ confirm progressive rates over time.
- Whole genome sequencing (WGS) and whole exome sequencing (WES) were successfully demonstrated at 5, 10, 20, 30, 50 and 100X coverage from upper arm capillary whole blood collections.
- BAM, BCL, FASTQ, VCF and additional related derivatives were successfully achieved with passing QC across all coverage ranges.
- A comparison of both standard and rare germline variation within the entire human genome set of diverse genes commonly impacts mutational processes in somatic cells.
- Given that somatic mutations are components of the biological and cellular ageing process, tracking of the changes in somatic mutations (i.e. SMR) demonstrates the rates and types of mutations across individuals and populations. This can be used to determine disease progression.
- Associations between deleterious variants and somatic mutations within longitudinal timepoints suggest evidence of gene interactions that drive mutagenesis.
- The SMR is used to identify the variant rate of change and longitudinal disease signatures within individuals by implementing standardized PICARD analysis across every longitudinal timepoint within the TimepointDx LCM™ analysis.
See White Paper for Details.
